
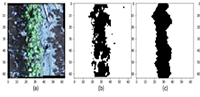
Resistance of complex interspecific cotton hybrids to wilt
Vol 5, Issue 2, 2024
VIEWS - 3764 (Abstract)
Download PDF
Abstract
In the article, the results of determining the degree of tolerance to wilt disease in the wilt background of hybrid families obtained using the method of interspecies complex hybridization in the cotton plant are analyzed. In the F2–F4 generations of interspecies complex hybrids, a large-scale process of separation according to the sign of tolerance to wilt disease takes place, and among the hybrids, plants that are not damaged by wilt disease, partially damaged plants, and severely damaged plants are separated. With this in mind, it was confirmed that it is possible to isolate lines and varieties with different levels of wilt damage from complex hybrid combinations of interspecies with different genomes by purposefully conducting selection work by the breeder. A number of wilt disease-tolerant families compared to control cultivars have been isolated and are recommended for use as a starting source in genetic selection studies.
Keywords
References
- Abdelraheem A, Zhu Y, Zhang J. Quantitative Trait Locus Mapping for Fusarium Wilt Race 4 Resistance in a Recombinant Inbred Line Population of Pima Cotton (Gossypium Barbadense). Pathogens. 2022; 11(10): 1143. doi: 10.3390/pathogens11101143
- Abdullaev AA, Salakhutdinov IB, Egamberdiev SSH, et al. Analyses of Fusarium wilt race 3 resistance in Upland cotton (Gossypium hirsutum L). Genetica. 2015; 143(3): 385–392. doi: 10.1007/s10709-015-9837-2
- Abdurakhmonov IY, Abdukarimov A. Application of Association Mapping to Understanding the Genetic Diversity of Plant Germplasm Resources. International Journal of Plant Genomics. 2008; 2008: 1–18. doi: 10.1155/2008/574927
- Abdurakhmonov IY. Cotton genomics and transgenomics in Uzbekistan. In: Proceedings of the International Cotton Genome Initiative Research Conference; 9–13 October 2012; Raleigh, NC, USA. pp. 1133–1142.
- Abdullaev AA. Cotton introduction in Uzbekistan—History and perspectives of using of plant introduction problems and perspectives. In: Proceedings of 4th National scientific-applied conference; 3–4 July 2009; Tashkent, Uzbekistan. pp. 59–61.
- Abdullaev A. Cotton Germplasm Collection of Uzbekistan. The Asian and Australasian Journal of Plant Science and Biotechnology. 2013; 7: 1–15.
- Abdurakhmonov IY. Evaluation of G. hirsutum exotic accessions from Uzbek cotton germplasm collection for further molecular mapping purposes. In: Proceedings of 2004 Beltwide Cotton Conference; 5–9 January 2004; San Antonio, Texas, USA. pp. 1133–1142.
- Kappelman AJ. Fusarium wilt resistance in commercial cotton varieties. Plant Disease Reporter. 1971; 52: 896–899.
- Kappelman AJ. Fusarium wilt resistance in cotton (Gossypium hirsutum). Plant Disease Reporter. 1975; 59: 803–805.
- Smith SN, DeVay JE, Hsieh WH, et al. Soil-borne populations of Fusarium oxysporum f. sp. vasinfectum, a cotton wilt fungus in California fields. Mycologia. 2001; 93(4): 737–743. doi: 10.1080/00275514.2001.12063205
- Solovyov AI. Wilt of Cotton [PhD thesis]. State Agricultural Publishing House of the UzSSR; 1954. pp. 18–22.
- Sidorova SF. Cotton wilting. The spread of agricultural diseases in the USSR in 1968–1972. MINISTER. 1973; 61–66.
- Marupov A. Fusarium disease of cotton. Agrarian Science. 2012; 22: 141–152.
- Allen SJ. The effects of crop rotation with wheat on the incidence of Fusarium wilt of cotton in Australia. In: Proceedings of the 2006 Beltwide Cotton Conferences; 3–6 January 2006; San Antonio, Texas, USA. pp. 81–84.
- Allen SJ. Eliminating seed-borne inoculum of Fusarium oxysporum f. sp. vasinfectum in cotton. In: Proceedings of the 2001 Beltwide Cotton Conferences; 9–13 January 2001; Anaheim, CA, USA. pp. 139–140.
- Allen SJ. Integrated disease management for Fusarium wilt of cotton in Australia. In: Proceedings of the World Cotton Research Conference; 13 September 2007; Narrabri, Australia. pp. 511–514.
- Allen SJ. Delayed planting as a control strategy for Fusarium wilt of cotton in Australia. In: Proceedings of the 2005 Beltwide Cotton Conferences; 4–7 January 2005; New Orleans, Louisiana, USA. pp. 131–135.
- Scheikowski L. Inoculation of rotation crops with Fusarium oxysporum f. sp. vasinfectum. In: Proceedings of the Beltwide Cotton Conferences; 4–7 January 2010; New Orleans, LA, USA. pp. 4–7.
- Smith SN. Persistence of Fusarium oxysporum f. sp. vasinfectum in Fields in the Absence of Cotton. Phytopathology. 1975; 65(2): 190. doi: 10.1094/phyto-65-190
- Wang B, Dale ML, Kochman JK, et al. Variations in soil population of Fusarium oxysporum f. sp. vasinfectum as influenced by fertilizer application and growth of different crops. Australasian Plant Pathology. 1999; 28(2): 174. doi: 10.1071/ap99029
- Kaul S. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000; 408(6814): 796–815. doi: 10.1038/35048692
- Armstrong GM. American, Egyptian, and Indian cotton-wilt fusaria their pathogenicity and relationship to other wilt fusaria. US Dep Agric Tech Bull. 1960; 1219: 1–19.
- Bennett RS. Method for rapid production of Fusarium oxysporum f. sp. vasinfectum chlamydospores. The Journal of Cotton Science. 2013; 17: 52–59.
- Bennett RS. Sequence characterization of race 4-like isolates of Fusarium oxysporum from Alabama and Mississippi. The Journal of Cotton Science. 2013; 17: 125–130.
- Li CQ, Liu GS, Zhao HH, et al. Marker-assisted selection of Verticillium wilt resistance in progeny populations of upland cotton derived from mass selection-mass crossing. Euphytica. 2013; 191(3): 469–480. doi: 10.1007/s10681-013-0917-z
- Egamberdiev SS, Ulloa M, Saha S, et al. Molecular Characterization of Uzbekistan Isolates of Fusarium oxysporum f. sp. vasinfectum. Journal of Plant Science and Molecular Breeding. 2013; 2(1): 3. doi: 10.7243/2050-2389-2-3
- Hillocks RJ. Fusarium wilt. In: Hillocks RJ (editor). Cotton Diseases. CAB International, Wallingford; 1992. pp. 127–160.
- Kim Y, Hutmacher RB, Davis RM. Characterization of California Isolates of Fusarium oxysporum f. sp. vasinfectum. Plant Disease. 2005; 89(4): 366–372. doi: 10.1094/pd-89-0366
- Wang B, Brubaker CL, Summerell BA, et al. Local origin of two vegetative compatibility groups of Fusarium oxysporumf. sp. vasinfectum in Australia. Evolutionary Applications. 2010; 3(5–6): 505–524. doi: 10.1111/j.1752-4571.2010.00139.x
- Wang B, Dale ML, Kochman JK, et al. Effects of plant residue, soil characteristics, cotton cultivars and other crops on fusarium wilt of cotton in Australia. Australian Journal of Experimental Agriculture. 1999; 39(2): 203. doi: 10.1071/ea98083
- Boboev SG. Muratov G. Interspecies hybridization of cotton. Eurasian Union of Scientists (ESU); 2020.
- Mirakhmedov SM. Development of a method for breeding wilt-resistant cotton varieties and their introduction into production V.sb. Cotton genetics. MINISTER Tashkent, Uzbekistan; 1972. pp. 89–97.
- Namozov SE, Egamberdiev A, Siddikov A. Inheritance of some horticultural traits in simple and double hybrids of cotton. Bulletin of Uzbekistan Agrarian Science. 2001; 6(4): 55–58.
- Narayanan SS, Santhanam V, Intergenomic hybridization in Gossypium for the improvement of cultivated allotetraploid cottons. In: Proceedings of the Fourth International Cotton Genome Initiative (ICGI) Workshop; 10–13 October 2004; Hyderabad, India. pp. 113–114.
- Cronn RC, Small RL, Haselkorn T, et al. Rapid diversification of the cotton genus (Gossypium: Malvaceae) revealed by analysis of sixteen nuclear and chloroplast genes. American Journal of Botany. 2002; 89(4): 707–725. doi: 10.3732/ajb.89.4.707.
- Saha S, Raska DA, Stelly DM. Upland Cotton (Gossypium hirsutum L.) x Hawaiian Cotton (G. tomentosum Nutt. ex Seem.) F1 Hybrid Hypoaneuploid Chromosome Substitution Series. Journal of Cotton Science. 2006; 10(3): 263–272.
- Dospehov BA. Methodology of field experience with the basics of statistical processing of research results, 5th ed. Rev. and add M.: Agropromizdat; 1985. p. 416.
Supporting Agencies
Copyright (c) 2024 Gulnoza Toshpulatova, Sayfulla Boboyev, Shaxzoda Muxammadiyeva
License URL: https://creativecommons.org/licenses/by/4.0/

This site is licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0).

Prof. Zhengjun Qiu
Zhejiang University, China

Cheng Sun
Academician of World Academy of Productivity Science; Executive Chairman, World Confederation of Productivity Science China Chapter, China
Indexing & Archiving
In the realm of modern agriculture, the integration of cutting-edge technologies is revolutionizing the way we approach sustainable farming practices. A recent study published in Advances in Modern Agriculture titled "Classification of cotton water stress using convolutional neural networks and UAV-based RGB imagery" has garnered significant attention for its innovative approach to precision irrigation management. Conducted by researchers from Institute of Data Science and the AgriLife Research and Extension Center of Texas A&M University (authors's information is below). This study introduces a novel method for classifying cotton water stress using unmanned aerial vehicles (UAVs) and convolutional neural networks (CNNs), offering a powerful solution for optimizing water use in agriculture.
Modern agricultural technology is evolving rapidly, with scientists collaborating with leading agricultural enterprises to develop intelligent management practices. These practices utilize advanced systems that provide tailored fertilization and treatment options for large-scale land management.
This journal values human initiative and intelligence, and the employment of AI technologies to write papers that replace the human mind is expressly prohibited. When there is a suspicious submission that uses AI tools to quickly piece together and generate research results, the editorial board of the journal will reject the article, and all journals under the publisher's umbrella will prohibit all authors from submitting their articles.
Readers and authors are asked to exercise caution and strictly adhere to the journal's policy regarding the usage of Artificial Intelligence Generated Content (AIGC) tools.
Asia Pacific Academy of Science Pte. Ltd. (APACSCI) specializes in international journal publishing. APACSCI adopts the open access publishing model and provides an important communication bridge for academic groups whose interest fields include engineering, technology, medicine, computer, mathematics, agriculture and forestry, and environment.



.jpg)
.jpg)

.jpg)
