
Asia Pacific Academy of Science Pte. Ltd. (APACSCI) specializes in international journal publishing. APACSCI adopts the open access publishing model and provides an important communication bridge for academic groups whose interest fields include engineering, technology, medicine, computer, mathematics, agriculture and forestry, and environment.
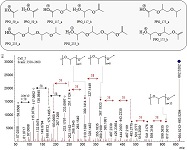
The (partial) replacement of synthetic polymers with bioplastics is due to increased production of conventional packaging plastics causing for severe environmental pollution with plastics waste. The bioplastics, however, represent complex mixtures of known and unknown (bio)polymers, fillers, plasticizers, stabilizers, flame retardant, pigments, antioxidants, hydrophobic polymers such as poly(lactic acid), polyethylene, polyesters, glycol, or poly(butylene succinate), and little is known of their chemical safety for both the environment and the human health. Polymerization reactions of bioplastics can produce no intentionally added chemicals to the bulk material, which could be toxic, as well. When polymers are used to food packing, then the latter chemicals could also migrate from the polymer to food. This fact compromises the safety for consumers, as well. The scarce data on chemical safety of bioplastics makes a gap in knowledge of their toxicity to humans and environment. Thus, development of exact analytical protocols for determining chemicals of bioplastics in environmental and food samples as well as packing polymers can only provide warrant for reliable conclusive evidence of their safety for both the human health and the environment. The task is compulsory according to legislation Directives valid to environmental protection, food control, and assessment of the risk to human health. The quantitative and structural determination of analytes is primary research task of analysis of polymers. The methods of mass spectrometry are fruitfully used for these purposes. Methodological development of exact analytical mass spectrometric tools for reliable structural analysis of bioplastics only guarantees their safety, efficacy, and quality to both humans and environment. This study, first, highlights innovative stochastic dynamics equations processing exactly mass spectrometric measurands and, thus, producing exact analyte quantification and 3D molecular and electronic structural analyses. There are determined synthetic polymers such as poly(ethylenglycol), poly(propylene glycol), and polyisoprene as well as biopolymers in bags for foodstuffs made from renewable cellulose and starch, and containing, in total within the 20,416–17,495 chemicals per sample of the composite biopolymers. Advantages of complementary employment in mass spectrometric methods and Fourier transform infrared spectroscopy is highlighted. The study utilizes ultra-high resolution electrospray ionization mass spectrometric and Fourier transform infrared spectroscopic data on biodegradable plastics bags for foodstuffs; high accuracy quantum chemical static methods, molecular dynamics; and chemometrics. There is achieved method performance |r| = 0.99981 determining poly(propylene glycol) in bag for foodstuff containing 20,416 species and using stochastic dynamics mass spectrometric formulas. The results highlight their great capability and applicability to the analytical science as well as relevance to both the fundamental research and to the industry.
Bifidobacteria serve as indicators of fecal contamination in tropical water bodies
Vol 1, Issue 1, 2020
Download PDF
Abstract
Human fecal contamination poses a significant public health concern in water sources, yet standard indicator microorganisms for detecting such contamination fail to pinpoint the exact source. The genus Bifidobacterium, particularly species like B. adolescentis and B. dentium, has been suggested as a potential marker for identifying human fecal pollution, though this proposal has yet to be tested in tropical settings. This study aimed to assess the presence of bifidobacteria in a water sample from the Mesolandia swamp in the Colombian Caribbean, as well as in 260 human fecal samples and 94 samples from domestic animals in a nearby settlement. DNA was extracted from each sample and subjected to PCR amplification with gender-specific primers targeting the 16S rRNA gene, followed by DGGE (Denaturing Gradient Gel Electrophoresis) separation. DGGE bands were then excised, re-amplified, sequenced, and compared to the GenBank database. The DGGE profiling revealed the presence of eight Bifidobacteria species in the water sample, matching those found in human feces. The proposed markers B. adolescentis and B. dentium were also detected in domestic animal feces. Despite the efforts, the study was unable to identify a unique Bifidobacteria species that could serve as a reliable marker for human fecal contamination in tropical environments under the evaluated conditions. Nevertheless, the methodology employed provided a more precise approximation to the source of fecal contamination than traditional cultural methods, as identical DNA sequences were found in both water and fecal samples.
Keywords
References
- Levengood JM, Hörman A, Hänninen M, et al. Water Security in a Changing World. Beyond One Health. 2018: 91-115. doi: 10.1002/9781119194521.ch4
- Rochelle-Newall E, Nguyen TMH, Le TPQ, et al. A short review of fecal indicator bacteria in tropical aquatic ecosystems: knowledge gaps and future directions. Frontiers in Microbiology. 2015; 6. doi: 10.3389/fmicb.2015.00308
- Waso M, Ndlovu T, Dobrowsky PH, et al. Presence of microbial and chemical source tracking markers in roof-harvested rainwater and catchment systems for the detection of fecal contamination. Environmental Science and Pollution Research. 2016; 23(17): 16987-17001. doi: 10.1007/s11356-016-6895-7
- Hellmér M, Stranddorf K, Seidel M, et al. Viral indicators for fecal contamination-a one-year viral metagenomic study of treatment efficiency in Danish waste water treatment plants. Danish Microbiological Society Annual Congress 2017. American Society for Microbiology, Copenhagen; 2017.
- Mayer RE, Reischer GH, Ixenmaier SK, et al. Global Distribution of Human-Associated Fecal Genetic Markers in Reference Samples from Six Continents. Environmental Science & Technology. 2018; 52(9): 5076-5084. doi: 10.1021/acs.est.7b04438
- Ballesté E, Bonjoch X, Belanche LA, Blanch AR. Molecular indicators used in the development of predictive models for microbial source tracking. Applied and Environmental Microbiology. 2010; 76(6): 1789-1795. doi: 10.1128∕AEM.02350-09
- Casanovas-Massana A, Gómez-Doñate M, Sánchez D, et al. Predicting fecal sources in waters with diverse pollution loads using general and molecular host-specific indicators and applying machine learning methods. Journal of Environmental Management. 2015; 151: 317-325. doi: 10.1016/j.jenvman.2015.01.002
- Gómez-Doñate M, Ballesté E, Muniesa M, et al. New Molecular Quantitative PCR Assay for Detection of Host-Specific Bifidobacteriaceae Suitable for Microbial Source Tracking. Applied and Environmental Microbiology. 2012; 78(16): 5788-5795. doi: 10.1128/aem.00895-12
- Regional Autonomous Corporation of Atlántico. Physicochemical characterization of wastewater discharges to water bodies in the Department of Atlántico and monitoring of the quality and current status of water sources in the department year 2011. Available online: http://www.crautonoma.gov.co/documentos/pomcas/RecursoHidrico/INFORME FINAL.pdf (accessed on 3 May 2020).
- Institute of Hydrology, Meteorology and Environmental Studies IDEAM. Guide for monitoring of discharges, surface and ground waters. Available online: http://corponor.gov.co/corponor/sigescor2010/TRAMITESYSERVICIOS/Guia_monitoreo_IDE AM.pdf (accessed on 3 May 2020).
- National Institute of Health (INS). Instruction manual for the collection, preservation and transport of drinking water samples for laboratory analysis. Available online: http://documentacion.ideam.gov.co/cgi-bin/koha/opac-detail.pl?biblionumber=11214&shelfbrowse_itemnumber=11791 (accessed on 3 May 2020).
- Rice EW, Baird RB, Eaton AD, Clesceri LS. Standard Methods for the Examination of Water and Wastewater (23rd Edition). Washington, D.C., USA: American Public Health Association. American Water Works Association, Water Environment Federation; 2017.
- Satokari RM, Vaughan EE, Akkermans ADL, et al. Bifidobacterial Diversity in Human Feces Detected by Genus-Specific PCR and Denaturing Gradient Gel Electrophoresis. Applied and Environmental Microbiology. 2001; 67(2): 504-513. doi: 10.1128/aem.67.2.504-513.2001
- Bonjoch X, Balleste E, Blanch AR. Multiplex PCR with 16S rRNA gene-targeted primers of Bifidobacterium spp. to identify sources of fecal pollution. Applied and Environmental Microbiology. 2004; 70(5): 3171-3175. doi: 10.1128∕AEM.70.5.3171-3175.2004
- Presidency of the Republic of Colombia. Decree 1594 of 1984. which partially regulates the use of water and liquid waste. Updated in National Decree 3930 of 2010. Available online: http://www.alcaldiabogota.gov.co/sisjur/normas/Norma1.jsp?i=18617 (accessed on 3 May 2020).
- Garcés-Ordóñez O, Vivas-Aguas LJ, Martínez M, et al. Diagnosis and evaluation of the quality of marine and coastal waters of the Colombian Caribbean and Pacific. Invemar Periodic Publication Series No. 4. Monitoring network for the conservation and protection of marine and coastal waters of Colombia (REDCAM). (Technical Report 2015). Santa Marta, Colombia: INVEMAR, MADS and coastal CAR; 2016.
- Al-Gheethi A, Ismail N, Efaq AN, et al. Solar disinfection and lime stabilization processes for reduction of pathogenic bacteria in sewage effluents and biosolids for agricultural purposes in Yemen. Journal of Water Reuse and Desalination. 2015; 5(3): 419-429. doi: 10.2166∕wrd.2015.074
- Burkhardt W, Calci KR, Watkins WD, et al. Inactivation of indicator microorganisms in estuarine waters. Water Research. 2000; 34: 2207-2214.
- Cabrala AC, StarkcHedda JS, Kolmb K, Martins C. An integrated evaluation of some faecal indicator bacteria (FIB) and chemical markers as potential tools for monitoring sewage contamination in subtropical estuaries. Environmental Pollution. 2018; 235:739-749. doi: 10.1016∕j.envpol.2017.12.109
- Rochelle-Newall E, Nguyen TMH, Le TPQ, et al. A short review of fecal indicator bacteria in tropical aquatic ecosystems: knowledge gaps and future directions. Frontiers in Microbiology. 2015; 6. doi: 10.3389/fmicb.2015.00308
- Flint HJ, Duncan SH, Louis P. The impact of nutrition on intestinal bacterial communities. Current Opinion in Microbiology. 2017; 38: 59-65. doi: 10.1016/j.mib.2017.04.005
- Korpela K. Diet, Microbiota, and Metabolic Health: Trade-Off Between Saccharolytic and Proteolytic Fermentation. Annual Review of Food Science and Technology. 2018; 9(1): 65-84. doi: 10.1146/annurev-food-030117-012830
- Gavini F, Delcenserie V, Kopeinig K, et al. Bifidobacterium Species Isolated from Animal Feces and from Beef and Pork Meat. Journal of Food Protection. 2006; 69(4): 871-877. doi: 10.4315/0362-028x-69.4.871
- Sabbioni A, Ferrario C, Milani C, et al. Modulation of the Bifidobacterial Communities of the Dog Microbiota by Zeolite. Frontiers in Microbiology. 2016; 7. doi: 10.3389/fmicb.2016.01491
- Turroni F, Milani C, Douwe Van Sinderen C, Ventura M. Bifidobacteria: Ecology and Coevolution with the Host. In: The Bifidobacteria and Related Organisms, Biology, Taxonomy, Applications. Academic Press; 2018. pp. 213-220. doi: 10.1016/B978-0-12−805060-6.00012-0
Supporting Agencies
Copyright (c) 2020 Sarmiento-Rubiano Luz Adriana, Yina García, Marianella Suarez-Marenco, Vanesa Inés Hoyos Solana, Jimmy E Becerra
License URL: http://creativecommons.org/licenses/by/4.0/

This site is licensed under a Creative Commons Attribution 4.0 International License (CC BY 4.0).
.jpg)
Beijing University of Technology, China



